idMotif: An Interactive Motif Identification in Protein Sequences, IEEE CGA, 2023, Link
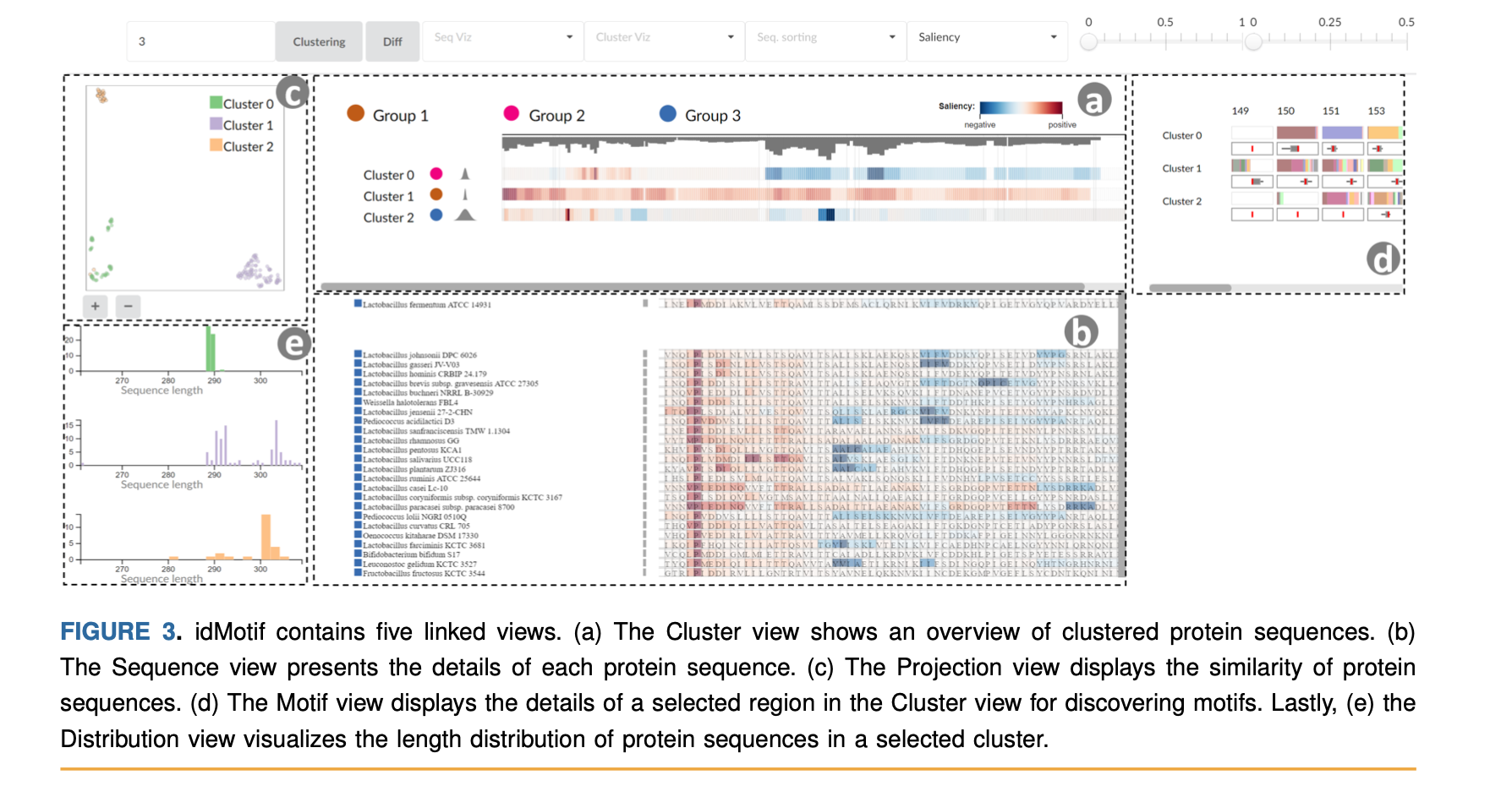
This article presents a visual analytics framework, idMotif, to support domain experts in identifying motifs in protein sequences. A motif is a short sequence of amino acids usually associated with distinct functions of a protein, and identifying similar motifs in protein sequences helps to predict certain types of disease or infection. idMotif can be used to explore, analyze, and visualize such motifs in protein sequences. We introduce a deep-learning-based method for grouping sequences and allow users to discover motif candidates of protein groups based on local explanations of the decision of a deep-learning model. idMotif provides several interactive linked views for between and within protein cluster/group and sequence analysis. Through a case study and experts’ feedback, we demonstrate how the framework helps domain experts analyze protein sequences and motif identification.
PepProEx Peptide and Protein Exploration Framework, IEEE Vis, 2023, Link
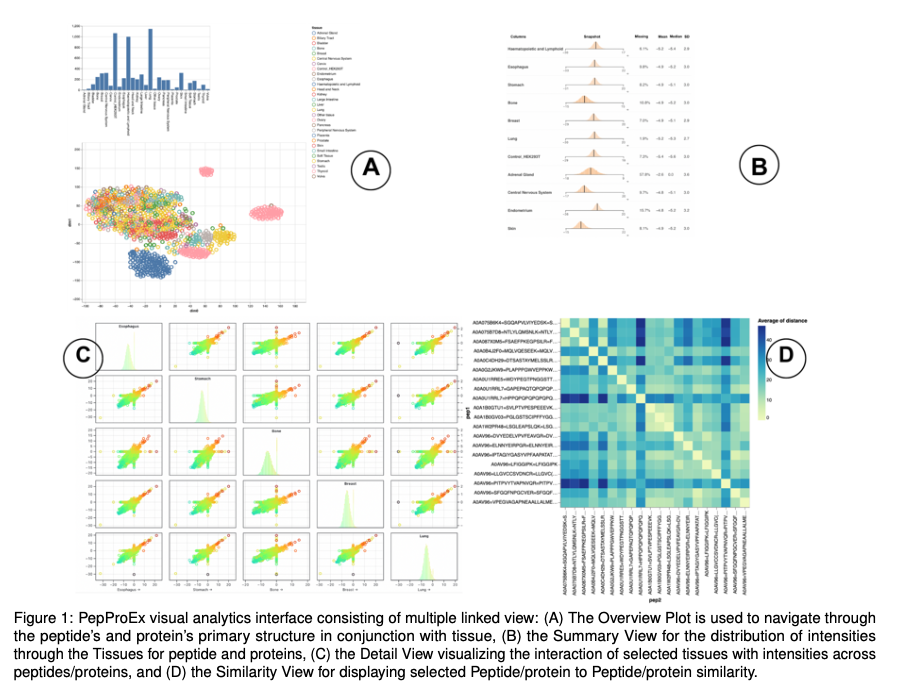
A VA framework, for unraveling peptide and protein interactions. In the framework, users can understand holistic trends, discover patterns, and get insights from the peptide/protein intensity data. Our framework provides multiple linked views to navigate intricate tissue/cancer-related dynamics. We enable researchers to shift effortlessly from an overview of the input data to granular statistical insights.
Data Processing and Classification, US Patent 2021-0182659 A1, Link
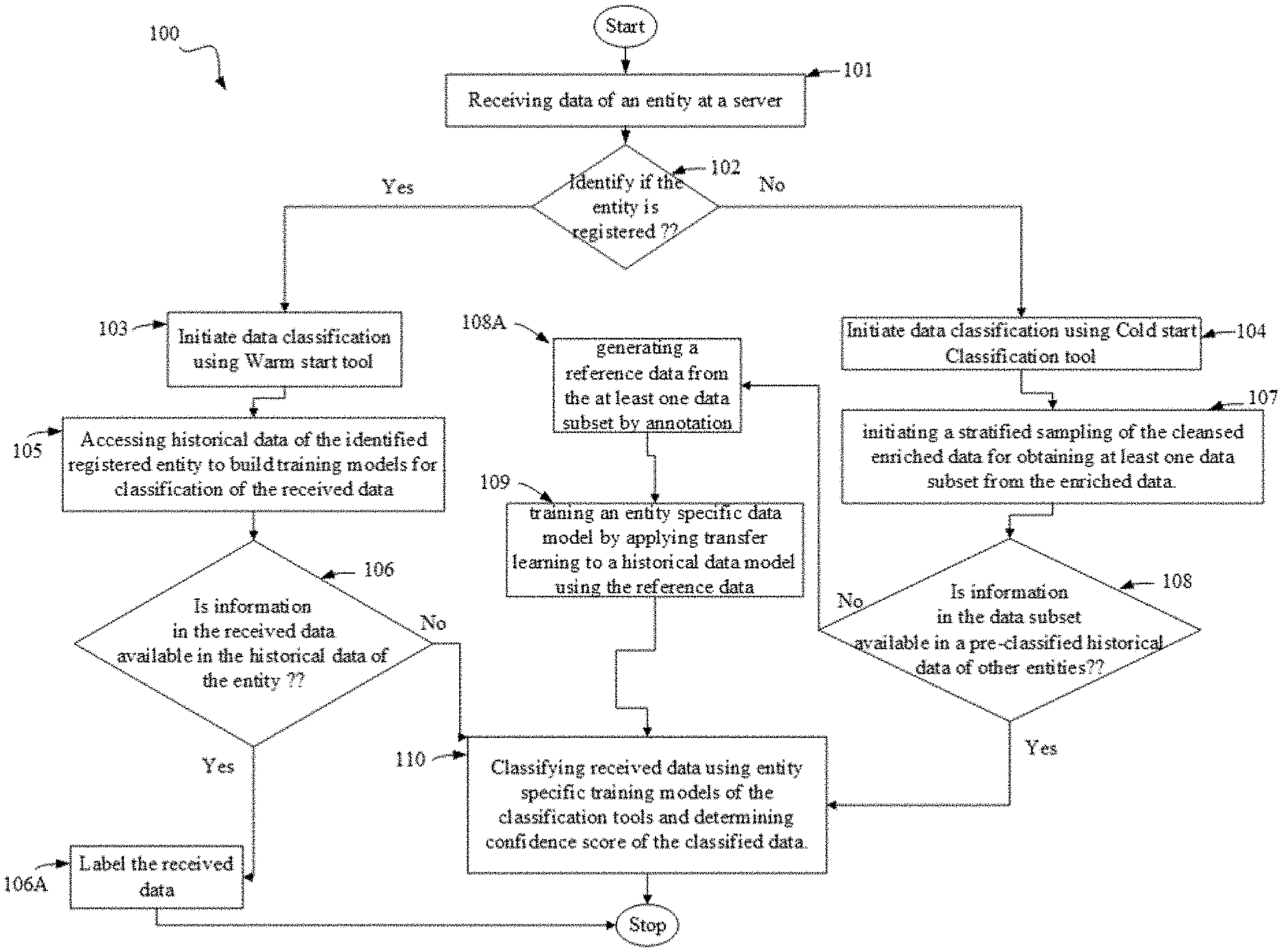
The present invention discloses a method, a system and a computer program product for data processing and classification. The invention provides warm start and cold start classification tools for classification of data obtained from known or unknown entities. The system and method are also configured to be employed over blockchain based networks.